Publications

NASH
Non-Alcoholic SteatoHepatitis, a liver disease evolving to CKD
Ratziu et al. (2022)
Hepatic and renal improvements with FXR agonist Vonafexor in individuals with suspected fibrotic NASH. Journal of Hepatology. 78 (3), 479-492.
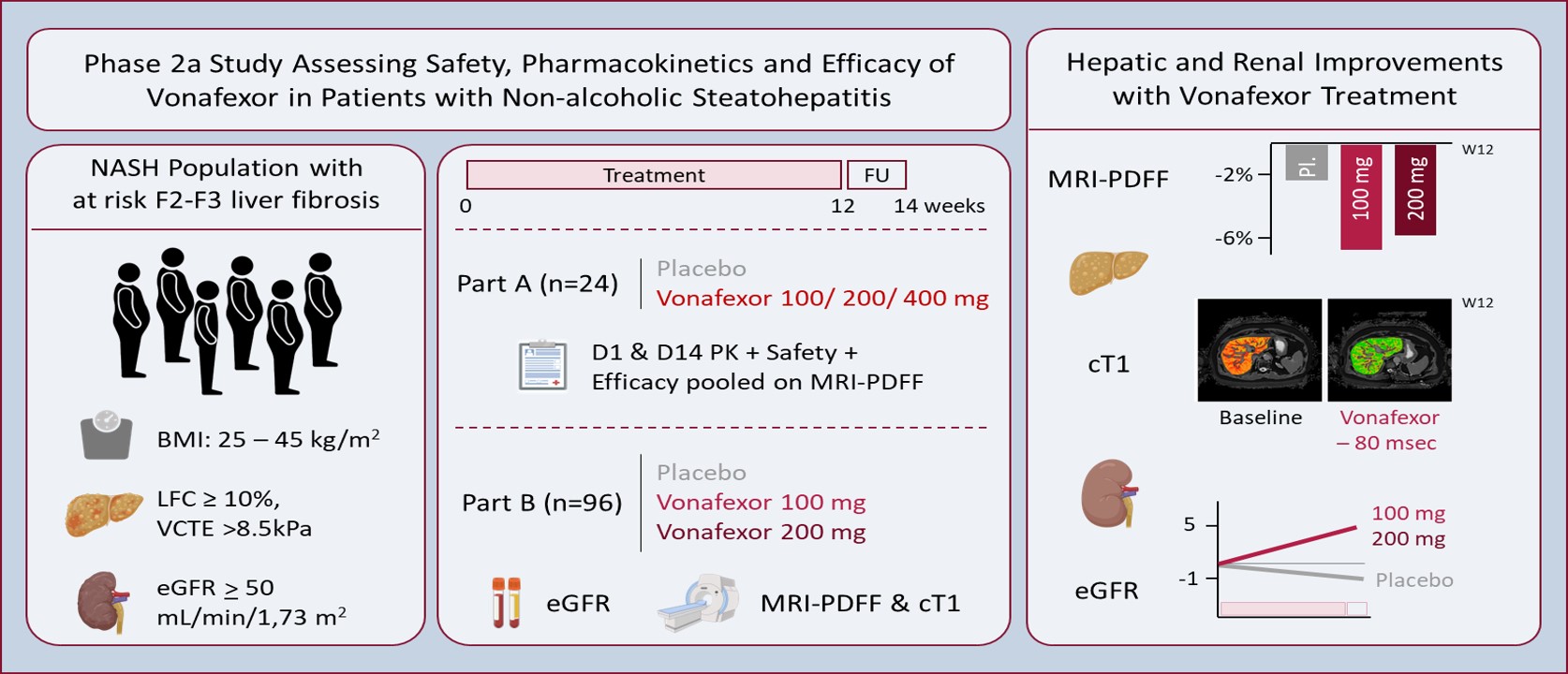
Non-alcoholic steatohepatitis (NASH) has become a leading cause of chronic liver disease worldwide. Affected patients are also at higher risk of developing chronic kidney disease. There are no approved therapies and only few options to treat this population. The phase IIa LIVIFY trial results show that single daily administration of oral vonafexor, an FXR agonist, leads in the short term to a reduction in liver fat, liver enzymes, fibrosis biomarkers, body weight and abdominal circumference, and a possible improvement in kidney function, while possible mild moderate pruritus (a peripheral FXR class effect) and an LDL cholesterol increase are manageable with lower doses and statins. These results support exploration in longer and larger trials, with the aim of addressing the unmet medical need in NASH.
Download full version Ratziu et al. (2022)
Other publications
– HBV
– Drug discovery platform
 Latest Press Release
Latest Press Release 